Abstract
This paper extends the numerical tuning of tree constants in genetic programming (GP) to the multiobjective domain. Using ten real-world benchmark regression datasets and employing Bayesian comparison procedures, we first consider the effects of feature standardization (without constant tuning) and conclude that standardization generally produces lower test errors, but, contrary to other recently published work, we find much less clear trend for tree sizes. In addition, we consider the effects of constant tuning – with and without feature standardization – and observe that (1) constant tuning invariably improves test error, and (2) usually decreases tree size. Combined with standardization, constant tuning produces the best test error results; tree sizes, however, are increased. We also examine the effects of applying constant tuning only once at the end a conventional GP run which turns out to be surprisingly promising. Finally, we consider the merits of using numerical procedures to tune tree constants and observe that for around half the datasets evolutionary search alone is superior whereas for the remaining half, parameter tuning is superior. We identify a number of open research questions that arise from this work.
Similar content being viewed by others
1 Introduction
Traditionally, the empirical modeling of data proceeds by a human analyst selecting models from some family (or families), and then optimizing a given model’s parameters, typically using a maximum likelihood formulation, to obtain a ‘best fit’ to the data; in the case of regression problems, this usually takes the form of minimizing a least-squares measure over a set of training data. Unless there is any good, a priori reason to adopt a specific model, the human analyst typically repeats this exercise for a range of models, and selects a final model using the candidates’ performance over a validation set disjoint to the training set. Finally, predictive performance is estimated over a test set that is disjoint to both the training and validation sets.
In practice, human analysts tend to consider only a limited range of potential data models since the process of parameter fitting and model comparison is tedious and time-consuming, a difficulty that has given rise to automated machine learning (AutoML) that seeks to mechanize this data fitting process [14] without relying on an expert analyst; in the context of the present paper, the genetic programming-based TPOT system [24] is noteworthy although it selects (albeit automatically) over a family of existing pre-processing and classification models rather than synthesizing entirely novel classifiers.
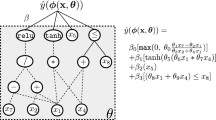
One of the promises of genetic programming (GP) is its ability to generate novel model structures driven by optimization of fitness over the dataset at hand rather than restricting the search for a data model to some prescribed set of candidates. In this context, the usual motivation of GP is slightly different from AutoML approaches although it shares the same objectives. It is widely considered, however, that while GP has the potential to synthesize data-driven model structures, optimization of that model’s parameters – the second part of the traditional, human-centered workflow – is a weak point that has received relatively limited attention in the GP community compared to areas like novel genetic operators, bloat, etc. More formally, a predictive model has the form of \({\hat{y}}=f({\mathbf {x}}; \varvec{\theta })\) where \({\hat{y}}\) is the model’s prediction, \({\mathbf {x}}\) is the vector of independent variables, and \(\varvec{\theta }\) is the real vector of model parameters. GP is well suited to searching over the space of functions \(f \in {\mathcal {F}}\), but is widely regarded as being poor at optimizing \(\varvec{\theta }\). Typical GP operations such as crossover and tree mutation are usually considered unlikely to determine optimal values of \(\varvec{\theta }\), for which sensitivity criteria may require close to the maximum available floating-point precision for good performance.Work on parameter tuning (also known as model calibration) in GP has been comprehensively reviewed in a recent paper by Kommenda et al. [17]. Most of the previous GP parameter tuning work has been carried out on regression problems, and this too is the focus of the present paper.
Kommenda et al. [17] used the well-known Levenberg-Marquardt (L-M) algorithm to optimize the constants in a GP tree with an algorithm that hybridized evolutionary and conventional numerical procedures to minimize the Pearson \(R^2\) correlation coefficient over a training set instead of the mean squared error metric that is more conventionally used in regression problems. Overall, Kommenda et al. concluded that conventional GP with nonlinear least-squares optimization of the constants was a (tied) top-ranking performer among a number of alternative GP and other models, hence we explore GP with constant tuning here. The present paper extends the work reported in [17].
On a closely related theme, Owen et al. [25] considered the effect of standardizing the features in GP to zero-mean/unit variance for a range of regression problems, a procedure widely used in other sub-areas of machine learning, but hitherto little-used in GP. These authors concluded that “the performance of GP can be greatly improved simply through z-score standardization of variables prior to training” with this “typically resulting in models that were smaller and generalized better than using unscaled variables”. These authors conjectured that placing all explanatory variables on the same scale made it easier for the GP search process to evolve appropriate values of tree constants. In a subsequent paper [7], the same authors extended their analysis to performing a single round of stochastic gradient optimization (1-SGD) on GP with and without feature standardization; we explore a wider combination of more extensive constant tuning in the present paper. In addition, Dick et al. [7] observed that GP with standardized inputs tended to evolve smaller trees, but once 1-SGD was included, standardization tended to produce trees larger than the baseline (unstandardized) GP. In the light of a number open issues identified in [7, 25], we therefore also consider the effects of dataset standardization alongside constant optimization, but extend this to the multiobjective framework.
The motivation of this paper has been to attempt to further align GP and traditional empirical data modeling, in particular, by exploring the factors involved in the parameter optimization phase of the modeling workflow described at the start of this section.
In terms of the original contributions of this paper:
-
We extend the results of [7, 25] on feature standardization to the multiobjective GP domain, and report its effects on testing error and tree size in Sect. 3.1. Extension to multiobjective GP stands in contrast to previous work on constant tuning that has, as far as we are aware, exclusively used single-objective GP.
-
We report a comprehensive exploration of the influence of optimizing the tree constants on the performance of GP models – both with and without feature standardization – by embedding the constant optimization inside the evolutionary loop which is presented in Sects. 3.2 and 3.3.
-
Most previous work on constant tuning in GP has embedded these optimization stages within the evolutionary loop. In Sect. 3.4 we revisit the effect of performing the constant optimization only at the end of a conventional GP run, something previously investigated (and dismissed) in very early work on constant tuning in GP [32].
-
We introduce a Bayesian statistical comparison of results; as far as we are aware, this is the first use of such methods in the GP literature.
Section 2 describes the methodology we have employed: the datasets, multiobjective genetic programming, formulation of the numerical optimization, and statistical testing. Section 3 presents the results of the paper, while Sect. 4 discusses the results’ implications and identifies possible future work and open research questions. Section 5 concludes the paper.
2 Methodology
2.1 Datasets
The datasets used in this study are shown in Table 1 having been used extensively in the past in the GP and wider machine learning literature. All bar one have been obtained from the UCI Repository [9]; the dow-chemical dataset was used as a competition at the EuroGP Conference in 2010.
For convenience, we have standardized all the predicted (i.e. dependent) variables for ease of comparison, what Owen et al. [25] termed partial standardization. The regressor (i.e. independent or feature) variables were either left unchanged or standardized, as described in particular experiments below. We have used the same, conventional procedure as in [25] to avoid bias by calculating any standardization transform over the training set, and applying the same transformation to both the validation and test sets. Both Kommenda et al. [17] and Owen et al. [25] used linear scaling of the predicted variable [15]. We have not employed a linear scaling procedure in the present work since Owen et al. [25] noted that it sometimes produces “erratic” predictive performance”.
We have adopted the standard machine learning procedure of partitioning each dataset into three disjoint subsets: a training set, a validation set, and test set [12]. The training set was used only for training after which the validation set was used to select a single, best-performing model from that GP run. Finally, the test partition was used to produce an independent estimate of generalization performance. This procedure is universal in mainstream machine learning, but some authors in the GP community omit the explicit model selection stage of using a validation set because they take the individual with the smallest training error as the selected model.
Two methods of analyzing cross-validation data simultaneously exist in the literature: the first performs a statistical test on either the mean (or median) test error over each fold for some number of independently initialized runs, while the second considers only the best-performing individual from each fold again over some number of independent runs. Both approaches are equally valid, but explore fundamentally different issues: the first explores ‘process’, namely the ability of, say, an evolutionary method or approach to produce good results ‘on average’. The second is an avowedly ‘engineering’ approach that aligns strongly with conventional machine learning practice, and focuses on the best obtainable model. We have adopted the first approach in this paper of comparing the mean test errors for each fold averaged over 30 repetitions since we are interested comparing methodologies. (In other settings, however, comparison of best-performing individuals may be more appropriate.)
We have used multiobjective GP (see Sect. 2.2) that produces a set of Pareto ‘equivalent’ models, and therefore we included an explicit model selection step. How to assign percentages of data to each of the three dataset partitions above is open to debate [12]: as an initial data pre-processing stage, we divided each dataset into ten folds for cross validation. To create each of the ten folds, 20% of the dataset was randomly sampled for testing. The remaining 80% was randomly divided into two sub-groups containing 70% and 10% of the total data, respectively; the former was used as the training set while the latter was used as the validation set. These are fairly commonly-used partitionings. Further, the sampling strategy employed meets the requirement of the statistical test employed – see Sect. 2.4.
2.2 Multiobjective genetic programming
The details of the multiobjective genetic programming (MOGP) algorithm used in this work are shown in Table 2; these parameter values have been used many times before by the present author. We have used a steady-state, as opposed to generational, evolutionary algorithm since our experience is that this give superior results – see Dou and Rockett [8] and the discussion therein for more details. Parents were selected for breeding by: i) sorting the population by Pareto rank allowing tied ranks, ii) mapping an individual’s rank to a scalar fitness with a linear function such that the best-ranked individuals got the largest fitness and the worst ranked zero fitness, and iii) selecting the parent stochastically biased by scalar fitness values – see [10, p.32] for full details.
2.3 Numerical optimization of the constants
Whereas Kommenda et al. [17] used the well-known Levenberg-Marquardt (L-M) algorithm to optimize the tree constants, we have chosen to use the Sequential Linear Quadratic Programming (SLSQP)Footnote 1 algorithm due to Kraft [18] for a number of reasons:
-
1.
Our previous experience, gained over a number of diverse application areas, is that the L-M algorithm either works very well and very quickly, or fails completely, an observation we suspect is due to the occasional pathological behavior of the approximation to the Hessian matrix of the objective function. Our experience of the SLSQP algorithm, which was designed for control applications, is that it is very robust, a property we consider important in the present application where the optimizer is to be embedded inside an evolutionary loop.
-
2.
Whereas the Levenberg-Marquardt algorithm can only minimize quadratic loss functions (i.e. the sum of squared residuals), SLSQP can minimize arbitrary functions requiring only the existence (but not explicit calculation) of at least the second derivatives of the loss functional. While not a factor in the present work on regression reported here, the use of SLSQP lays the foundations for future work on, for example, robust regression in the presence of outliers where more complex loss functions are generally required.
-
3.
SLSQP can also accommodate both equality and inequality constraints, which, again while not used in the present work, may be useful in future for constraining the constant values to, say, physically-meaningful values.
One of the requirements of both the L-M and SLSQP algorithms is the closed-form calculation of derivatives of the objective function w.r.t. the parameters to be optimized. In this paper, we have calculated the necessary derivatives using the automatic tree transformation rules described by Rockett et al. [30], which provide the exact value of the derivative (subject, of course, to normal rounding errors). Key to maintaining the differentiability of this automatic tree differentiation is replacing the commonly-used (protected) division GP operator with the analytic quotient (AQ) operator proposed by Ni et al. [21]. This has the advantage of guaranteeing that the function composition implemented by any GP tree is analytic, and therefore differentiable up to at least second order. Aside from the advantages of the AQ cited in [21], Nicolau and Agapitos [22] have recently reported that the AQ operator provides superior generalization over a range of regression problems compared to trees using protected division. Hence, given a tree for which we require to optimize the constant values, we can evaluate the necessary partial derivatives for SLSQP by: i) automatically generating the derivative trees of our target tree, and ii) evaluating those derivative trees by normal recursive tree traversals of the derivative trees for some given specific input. See [30] for further details of the automatic tree differentiation transformations.
Moreover, the work of Kommenda [17] used unprotected division which is manifestly not analytic when the denominator is zero, and for which derivatives do not therefore exist; how this has been handled was not discussed. Over and above attempts to evaluate derivatives at singular points, even protected division has the property that as x becomes very small but \(| x | > 0\), the quotient 1/x can become very large leading to numerical instabilities in the values predicted by the GP tree. We believe this is the underpinning reason for the conclusions of Nicolau and Agapitos [22] – see Ni et al. [21] for a longer discussion; such numerical instabilities also appear to have been evident in the results of Dick et al. [7] after (protected) division was included in their trees.
The setting of the convergence criteria for the SLSQP optimization algorithm requires some care since this may be embedded within an evolutionary optimizer, and ensuring robust performance is essential. The SLSQP routine was set to terminate when either: (1) the change in relative value of the objective (MSE) was > \(10^{-4}\), a criterion that ensures that the around a quarter of the most significant digits of an IEEE-754 compliant floating-point number were stable, or (2) the number of internal evaluations of the objective function exceeded 50. In practice, a significant number of trees would have required many more than 50 optimizer iterations to meet the convergence criterion of \(10^{-4}\) although terminating after 50 iterations invariably produced an approximate therefore useful although clearly not exactly converged solution. Our experience is that many of the generated trees appear poorly conditioned [13] resulting in very slow convergence. Terminating optimization of these trees after 50 iterations was thus a compromise between accuracy and excessive run time. The iteration limit of 50 was selected based on initial experimentation that resulted in around 80% of the offspring converging to the \(10^{-4}\) relative convergence limit within 50 iterations. (It is noteworthy that Kommenda et al. [17] used only a fixed number of 10 iterations for their L-M algorithm, and did not specify a convergence criterion on the objective.)
Formally, constant optimization can be formulated by considering a mapping implemented by a tree as \({{\hat{y}}} = f({\mathbf {x}}, \varvec{\theta })\), where \({\mathbf {x}}\) is the input vector of features, and \(\varvec{\theta }\) is the vector of (mutable) constants in the tree; \(y \in {\mathbb {R}}\), \({\mathbf {x}} \in {\mathbb {R}}^n\), and \(\varvec{\theta } \in {\mathbb {R}}^m\), where n, m are the numbers of input features and the number of mutable tree constants, respectively. For the purposes of constant optimization, we can regard the input vector \({\mathbf {x}}\) as a constantFootnote 2 (since, for a given data record, it is fixed by the training set), and the vector of parameters \(\varvec{\theta }\) as the variables in the minimization problem, the values of which can be optimized using the gradient-based SLSQP routine.
2.4 Statistical testing
Traditionally, the comparison of machine learning results has been carried out using a null hypothesis statistical test (NHST) [5]. NHSTs, however, have received considerable criticism – see, for example, Benavoli et al. [2] and references therein. In essence, an NHST calculates the ‘wrong’ sort of probability to be able to definitively judge differences between two methods: it calculates \(\Pr\)(data given an assumed null hypothesis) thereby requiring a contentious threshold (\(\alpha\)) on the calculated p-value in order to make a judgment on the statistical significance or otherwise of a discrepancy with the null hypothesis. Furthermore, a positive outcome from an NHST is commonly misinterpreted as somehow ‘proving’ superiority rather than being a highly conditioned inference on the supportability of a null hypothesis. In contrast, a Bayesian procedure that estimates \(\Pr (\text {a difference given the data})\) is what is really needed, and facilitates much more useful statements along the lines of “method A is superior to method B with a probability of x”. An NHST, on the other hand, only permits the much weaker statement that any differences between the methods are not consistent with the null hypothesis at the \((1-\alpha )\) confidence level. Consequently, in this work we have employed the Bayesian hypothesis testing procedures of Benavoli and co-workers [2]. In particular, we have used the Bayesian correlated t-test [3] to compare pairs of methods; the implementation used was taken from the baycomp Python packageFootnote 3.
Conventionally, the research question such as exploring the significance of the difference in mean errors over a cross-validation test has been addressed using a non-parametric test such as the Wilcoxon signed-rank test [5]. The motivation for using a signed-rank test as opposed to a parametric t-test (which generally exhibits greater statistical power) is that the ‘textbook’ t-test requires that the samples are independent; in cross-validation, of course, the samples are correlated because the training sets across different folds overlap [3]. In a frequentist setting, Nadeau and Bengio [20] proposed a correlated t-test that compensates for these correlations, a correction that was subsequently used by Corani and Benavoli [3] to devise a Bayesian correlated t-test. It is the Bayesian correlated t-test of Corani and Benavoli that we use here.
The other technical requirement of a t-test is that the samples are normally distributed, which can usually be ensured by averaging over some modest number of repetitions and appealing to the central limit theorem.
Interpretation of the Bayesian test requires some clarification: since it estimates the posterior probability of a difference (conditioned only on the data), the (somewhat arbitrary) NHST criterion of a “95% significance level” is of no relevance here. Rather, the Bayesian test returns a direct measure of belief in there being a difference between the two methods. There is, however, inevitably some element of subjectivity in interpreting posterior probability measures: for example, if the posterior probability of method A being superior to method B is 0.6, is this significant? In this work, we regard posterior value of 0.6 as providing weak evidence of superiority with larger values being progressively more persuasive.
To statistically compare two methods, we have performed 10-fold cross validation (CV) over each of the datasets in Sect. 2.1. We have adopted the protocol of repeating the training 30 times for each dataset fold each with different initial populations, and taking the individuals with the smallest validation set error from each of the 30 repetitions. The test error for each fold was then estimated by averaging the test set errors from the best validation error individual from each of the 30 repetitions. Ten mean test set errors were similarly calculated, one per fold, and then used in the Bayesian correlated t-test described above to calculate the posterior probability of difference between the two methods being compared.
The Bayesian correlated t-test [2] can be most easily understood by examining the posterior probability density functions (PDFs) of the measured differences between the two methods being compared. As an example, a posterior density (adapted from an image generated directly by the baycomp package) is shown in Fig. 1 for the comparison of two treatments.Footnote 4 The random variate plotted on the abscissa is the difference in the performances paired by fold for the two methods. From inspection of the plot in Fig. 1, it is clear that almost all the probability mass lies to the right of the origin meaning that the first compared method has a higher score than the other method with a posterior probability equal to the integrated PDF to the right of the origin. We re-emphasize that Fig. 1 depicts a posterior probability distribution computed from the Bayesian correlated t-test, and not a histogram of experimental results.
In addition to calculating the posterior probabilities for the test MSE results, we have also statistically compared the tree sizes of the best validation error individuals used for the MSE comparisons, again averaged over each of the 30 repetitions per fold. That is, the same individuals that were included in the MSE analysis were also included in the analysis of tree sizes using the same averaging methodology.
3 Results
We have performed an extensive series of statistical comparisons for various configurations, as detailed in the subsequent sub-sections. Throughout the present section, we adopt the following shorthand to allow us to concisely distinguish between the different experimental configurations:
-
We use “no tuning” to denote that the GP training has been done without any optimization of the constants (other than that produced by the evolutionary process).
-
The term “tuning during”, on the other hand, denotes that constant optimization has been performed on every child tree generated during evolution.
-
Finally, “tuning at end” is used to describe the situation where the final population has been evolved without any constant optimization during evolution, but the constants in every individual in the final population have been optimized just once at the end of the GP run.
Full numerical results of the Bayesian correlated t-tests under different conditions are shown for each of the ten datasets with the probability values rounded to 3 decimal places (d.p.) and the mean test MSE values to 4 d.p. Probabilities \(> 0.995\) are thus rounded to unity.
3.1 Baseline vs. baseline + standardization (‘No Tuning’)
We have run the MOGP algorithm on each of the ten partitions of the datasets for both the baseline (unstandardized) GP and the baseline GP with standardized features; in all cases, the predicted values were standardized for convenience, as explained above. The objective of this set of experiments was to explore the observations of Owen et al. [25] that standardization of the predictor variables improved generalization performance while reducing tree sizes. There was no constant optimization in any of the results described in this sub-section.
The comparisons between the results from the baseline GP and the baseline GP with standardized features are shown in Table 3. These results extend those in [25] and [7] to the steady-state multiobjective domain; in addition, they also add Bayesian statistical testing. For test MSE, the upper pair of numbers in each row gives the probabilities that each entry has a larger MSE than its comparator along with the the probability of the complementary event – these two probabilities sum to unity, of course; the lower pair of numbers in each row show the observed mean MSE values of each method over the ten data folds.
For example, in the case of the airfoil-self-noise dataset (first row of Table 3), the baseline GP has the larger test MSE with a probability 1.000, while the baseline GP with standardization has a larger MSE with a probability of 0.000. (These figures are the probability masses mentioned above and illustrated in Fig. 1a.) The mean MSE values immediately below record that the baseline GP had a mean test MSE of 0.5352 while the baseline plus standardization had a mean test MSE of 0.4264. Since smaller is better for MSE, we can conclude that the baseline GP + standardization produces a smaller (i.e. better) test MSE with a probability of 1.000 (i.e. certainty) for this particular dataset. Similarly, the third and fourth columns of Table 3 record the probabilities that the baseline GP with standardization produces larger trees and the (complementary) probability that the baseline GP produces larger trees – cf. Figure 1b. Again, to take the airfoil-self-noise dataset as an example, the probability that the baseline GP produces larger trees is 0.339 whereas there is a probability of 0.661 that standardization produces larger trees. We can conclude from the example of the airfoil-self-noise dataset that standardization results in larger trees with a probability of 0.661; the mean tree sizes are 143.0 for the baseline GP and 147.4 for baseline GP + standardization, respectively. The size effect in this case is, however, modest.
Taken overall, the results in Table 3 show that feature standardization generally produces lower values of MSE than the baseline algorithm, often with probabilities approximating absolute certainty (1.000 to 4 d.p.). The one deviation from the above trend is for the yacht-hydrodynamics dataset for which the baseline GP produces the lower MSE with a probability of 0.765.
As regards tree sizes, the trend in Table 3 is mixed. Standardization tends to produce smaller tree sizes for the servo and yacht-hydrodynamics, but larger trees for boston-housing and dow-chemical datasets. For the remaining datasets, the differences appear marginal, and in many cases, the size effects are small. For example, for the concrete-strength data, the mean node counts without and with standardization are 134.2 and 136.2, respectively; the statistical calculations, on the other hand, suggest that standardization yields larger trees with a probability of 0.643.
3.2 Effects of constant optimization on the baseline GP (‘Tuning During’)
The results of comparing the baseline GP with and without the constant optimization procedure described in Sect. 2.3 are shown in Table 4, namely, no tuning vs. tuning during. Here the constant optimization was applied to each child as soon as it was created and before it was inserted into the population, what we term ‘tuning during’ (evolution). The same interpretations of the results need to be applied here as in Sect. 3.1.
Viewed overall, we can observe from Table 4 that ‘tuning during’ (i.e. constant optimization of every child throughout the GP run) produces smaller MSE values compared to the baseline GP; most tests return a probabilities of 1.0 with the least favorable result for the qsar-aquatic-toxicity dataset with a probability of 0.918. The general trend in Table 4 agrees with the observations of Kommenda et al. [17] that constant optimization reduces MSE values although the present work extends their observations to the multiobjective GP domain.
For the tree size comparisons in Table 4, generally, tuning during evolution produces smaller trees (along with smaller MSE values). In some cases (airfoil-self-noise and energy-efficiency-cool) the mean tree sizes are around 36% smaller; in the case of the boston-housing dataset the difference is only 5% smaller. For the most part, the statistical tests suggest very strong evidence to support tuning producing smaller trees.
3.3 Constant optimization combined with feature standardization (‘Tuning during’)
From the preceding MSE results in Sect. 3.1, feature standardization produces generally better outcomes when compared to the baseline GP without constant tuning. An obvious combination is to explore if feature standardization followed by constant tuning during evolution can produce even better results; the procedure here is identical to that used for generating the the results in Table 4 except that the dataset features in both compared methods have been standardized – that is, we are comparing baseline GP + standardization vs. baseline GP + standardization + tuning during. If the conjecture of Dick et al. [7] that standardization produces trees in which the constants are easier to determine due to reduced ranges is valid then standardization may benefit the explicit constant optimization process by posing an ‘easier’ optimization task. The results from exploring this hypothesis are presented in Table 5.
As with the similar (but no-standardization) results in Table 4, the effect of constant optimization (tuning during evolution) produces consistently smaller MSE values; all datasets apart from the dow-chemical and qsar-aquatic-toxicity return emphatic results with posterior probabilities of unity. Even for these last two datasets, the probabilities of 0.785 and 0.788, respectively, are still fairly strong evidence for the superiority of tuning-during.
The comparison over node counts again indicates somewhat different behavior to that exhibited without standardization. Many of the statistical outcomes indicate marginal/no differences, and even where the posterior does offer clear evidence (e.g. auto-mpg), the size effects appear small.
Table 6 compares the results of the baseline GP followed by constant tuning during vs. the baseline GP + standardization + constant tuning during evolution. In other words, it explores the influence of adding feature standardization before constant tuning during evolution.
The clear picture that emerges from Table 6 is that standardization produces better MSE results. On the other hand, a comparison of nodes counts indicates that standardization generates larger trees. Taken together with the smaller test errors, this implies that standardization allows the evolutionary process (in conjunction with constant tuning) to find better-performing but more complex predictive models; this, of course, is an entirely acceptable trade-off in the the empirical modeling of data.
3.4 Constant optimisation after baseline GP (‘Tuning-at-end’)
One obvious question arising from this work is: does running constant optimization just once after the baseline GP produce comparable results to embedding the constant optimization within the evolutionary loop? The present section expands the work reported in [17], which did not consider constant tuning after evolution. Here we have tuned the constants in every individual in the final evolved population since we wanted to explore the extent of coupling between the search for the ‘best’ model structure (performed principally by the evolutionary search) and constant optimization (performed solely by the numerical optimization).
The results of these experiments for the baseline GP without feature standardization are shown in Table 7. The maximum number of iterations for tuning-at-end was set at 500 on the basis that this was – somewhat arbitrarily – ten times the limit used for tuning-during; initial experimentation also suggested that if an optimization had not converged after 500 iterations, it was not likely to converge in any feasible number of iterations. The iteration limit of 500 is thus another compromise between accuracy and run time.
The positive benefits for the test error of constant tuning are apparent for most datasets although the trend is reversed for dow-chemical, while qsar-aquatic-toxicity suggests no difference. The result for the dow-chemical dataset is interesting in that the mean test error is actually increased by constant tuning at the end. This implies that, in this case, tuning (of presumably over-parameterized models) has resulted in significant overfitting since there is no counter-balancing evolutionary pressure to reduce the size of the individuals. Why over-fitting seems to have happened for the dow-chemical dataset but not for the others is unclear.
The picture for the comparison of node counts for tuning-at-end is rather more mixed than for the MSE measure. For three datasets (concrete-strength, energy-efficiency-cool and energy-efficiency-heat), post-evolution tuning (tuning-at-end) produced smaller best-test individuals. For auto-mpg and probably dow-chemical, the mean node counts are unchanged by tuning after. For the remaining datasets, tuning-at-end selects larger trees. It is worth emphasizing that the pairs of populations considered here were structurally identical – the differences are only in the values embedded in the constant tree nodes and not the tree morphologies. It is thus interesting that in some cases, constant tuning selected smaller ‘best’ trees from the same populations, and in other cases, larger trees. It is also interesting that for the dow-chemical dataset trees of roughly the same sizes were selected as the best performers both before and after tuning-at-end, but the post-tuning results suggest significant overfitting.
3.5 Constant optimization after baseline GP with standardization (Tuning at end)
Table 8 summarizes similar results to Table 7 in the previous sub-section except here the features were standardized for both comparators. Turning to the MSE results in Table 8, the clear trend is of tuning -at-end producing superior results.
Unlike the corresponding results without standardization, there is a clear trend for node counts with tuning-at-end (+ standardization) producing statistically larger trees with the exception of the yacht-hydrodynamics dataset where there appears to be no difference.
3.6 Tuning-during vs. Tuning-at-end
Since tuning appears to offer some advantages – whether during or after evolution – at least for the MSE results, we have compared the baseline GP with ‘tuning-during’ evolution to ‘tuning-at-end’ over the final population; note standardization was not employed in this comparison. The objective here was to determine whether ‘tuning-during’ or ‘tuning-at-end’ was preferable. The results for this comparison are shown in Table 9.
For MSE, the picture presented in Table 9 is mixed. Tuning-at-the-end produces better test MSE figures for the energy-efficiency-cool and energy-efficiency-heat datasets, while for the remainder, tuning-during is the better option.
The conclusion for the node count comparisons is unambiguous: tuning-during produces smaller best-performing trees presumably because this method is able to impose an evolutionary pressure that simultaneously reduces both (training) MSE and tree size.
Repeating this analysis but for the case of standardized features produces the results in Table 10. Statistically, the trend is clear with tuning-during performing best across all ten datasets although for the airfoil-self-noise dataset, the difference in mean test errors is in the third decimal digit suggesting the that the size effect may not be too great. Comparisons over the numbers of node counts are also unambiguous with tuning-during producing smaller trees all with compelling probabilities and size effects of around 10%.
4 Discussion & future work
4.1 The effects of feature standardization on the baseline GP
Section 3.1 presented an analysis of the effects of feature standardization on both the test MSE and the tree sizes, which need to be set alongside those of Owen et al. [25] and Dick et al. [7]. Overall, and using a similar set of benchmark datasets, the MSE comparisons qualitatively agree with those of Owen at al. [25] in that feature standardization tends to reduce MSE although like [25] and the continuation work of Dick et al. [7], we too observe deviations from this trend. In particular, the yacht-hydrodynamics dataset shows evidence (\(\Pr\) = 0.765) that standardization degrades MSE. As with such cases observed in [7, 25], the reason is not clear. Even if the features in the yacht-hydrodynamics dataset were already (close to) standardized, a redundant application of the standardization procedure would have produced very little difference in performance rather than the observed and serious degradation. Establishing the reason why some datasets are degraded by standardization whereas some are improved requires further research. Similar off-trend results were noted elsewhere in Sect. 3, and we return to discussing those below.
Apart from the single case of qsar-aquatic-toxicity which returns no difference, the effects of feature standardization in Sect. 3.1 are that standardization can both increase tree size and decrease it depending on dataset. These results are thus at variance with what was seen in [7, 25] where standardization (alone) was observed to reduce tree size. The speculation in [25] was that standardization reduces the range of constants that a tree needs to synthesize in order to fit the data; indeed, in a study of pruning of baseline-type GP trees [29], we observed that significant numbers of individuals terminated with subtrees of the form of a constant combined with another constant using a binary operation, presumably serving to generate constants outside the range of those available in the a priori terminal set. Thus it is intuitively reasonable to suggest that standardization may reduce a tree’s need to synthesize ‘larger’ and ‘smaller’ constants leading to smaller overall tree sizes. This speculation is not, however, supported by the results in the present paper.
One possible explanation for the reason why Owen et al. observed decreases in tree sizes with standardization but we sometimes observe the opposite is that those researchers employed a generational, single-objective GP. The present work used a steady-state multiobjective GP in which model complexity was controlled only by (simultaneously) minimizing a node count measure within a Pareto framework. Although we observe some increases in trees sizes, this is accompanied by a (highly desirable) reduction in test MSE implying that standardization facilitates the evolution of better generalizing but more complex models. This is a perfectly acceptable trade-off in data modeling, and very distinct from tree bloat, which is the growth in tree size without accompanying improvement in (MSE) performance.
Reconciling the two diametrically opposed observations about the effect of standardization on tree size again requires further work, possibly focusing on the differences between generational and steady-state evolutionary approaches. In addition, careful reading of [7, 25] suggests there may be other methodological differences that will need to be systematically explored. For example, in [7], their equivalent of our baseline GP algorithm included protected division operators whereas their standardized version of the same algorithm did not.
Finally on this point, there is evidence in the literature [22, 23] that suggests the composition of the function set can affect both evolutionary search and generalization performance. Under standardization, regressors which may originally all have been non-negative can acquire negative values. One could speculate that in this scenario, the GP search has to generate negative constants from the positive constants available in the function set leading to an increase in tree size. To counter this, the function set used in the present work also included a unary minus node that can – in principle – generate a negative value very straightforwardly. Nonetheless, the interaction of the function set is another area for examination in future work.
4.2 Effects of constant optimization
In this sub-section, we discuss the effects of constant tuning across various configurations of GP since the effects are inter-related.
4.2.1 Baseline GP – Tuning during
For the baseline GP, Table 4 displays a very clear and unambiguous trend of constant optimization during evolution producing statistically smaller test MSE values. This conclusion agrees with the observations of Kommenda et al. [17] for fitting error – in their case, Pearson’s \(R^2\) coefficient – obtained using a single-objective paradigm. In addition, our tuning-during results simultaneously show much smaller trees were produced, again with large probabilities.
That constant tuning produces superior test set errors compared to the baseline GP is not surprising since tuning very directly minimizes the training error for a given tree structure. Within an evolutionary paradigm, there is clearly a pressure to produce smaller training MSE values, but balanced by the simultaneous pressure to produce ever smaller trees. Within a Pareto framework, the multiobjective GP typically produces a range of final models extending from small and underfitted through to large and overfitted from which a model exhibiting the best trade-off between bias and variance [11] can be selected using a validation set: see Sect. 2.
The clear trend from Table 4 is that tuning-during also tends to produce smaller, as well as better generalizing, models. Optimizing the constants for every child created during evolution in a Pareto framework will again tend to impose an evolutionary pressure that favors smaller trees. The fact that a tree does not have to synthesize ‘optimal’ constants by combining a number of fixed, constant leaves, but can use fewer, optimizable constants to achieve a lower MSE suggests that the evolved trees will be both smaller and have lower training error. It is very unlikely, however, that tuning-during removes all redundancies from the GP trees thereby reducing them to their minimum size for a given mapping. Nonetheless, it would be instructive to explore in further work if the trees produced with tuning-during had fewer instances of, for example, (redundant) constant-binary operation-constant terminations.
The present work also extends the results of Kommenda et al. [17], who did not directly compare tree sizes.
4.2.2 Baseline GP with standardization – Tuning during
Section 4.2.1 has discussed the beneficial results of tuning-during on the baseline GP without feature standardization. Since standardization (generally) improves the performance of the baseline GP in the absence of constant tuning, Sect. 3.5 explored whether including standardization prior to tuning-during provides additional benefits. Table 5 compares the baseline GP with standardization vs. the baseline GP with standardization + tuning-during. As with the results discussed in Sect. 4.2.1, standardization also produces statistically better test MSE values as well as generally smaller trees when compared to the baseline GP with standardization. For almost all datasets, however, tuning-during with standardization appears to exhibit a small size effects. So although tuning here has a clear beneficial effect on test MSE, the implication is that it is feature standardization alone that influences final tree size.
Table 6 compares the baseline GP + tuning during with the standardized baseline GP with tuning-during to measure the influence of feature standardization on the constant optimization process. The results are unambiguous: standardization improves test MSE while producing larger trees. In some cases, the size effects of the node count differences are quite large at \(\sim\)35%, while in others only around 5%. Quite why we observe a consistent increase in tree size with standardization is unclear. It is also worth further work to reconcile the results on tree size obtained here with the observation of Dick et al. [7] that a single iteration of stochastic gradient descent increased the sizes of their trees when using standardized datasets.
Up to this point, we can conclude that:
-
Feature standardization almost always benefits generalization but at the expense of creating larger models
-
Tuning the tree constants has a positive effect on generalization while reducing tree sizes relative to the same algorithm with no tuning.
-
The benefits of standardization are not universal, and there are some datasets that fall outside the trend: for example, standardization appears to produce inferior generalization for the yacht-hydrodynamics dataset in Table 3. As pointed out in [25], it would be helpful to understand what properties of these datasets lead to these outcomes.
4.2.3 Tuning during vs. Tuning at end
The motivation for exploring tuning-at-the-end of the GP was to try to further elucidate the influence of constant tuning. From the foregoing results, it is perhaps expected that performing even this more restricted form of constant optimization would improve test MSE results compared to the as-evolved population, and this indeed is the clear trend shown in Table 7 for the unstandardized baseline GP. This trend is the exact opposite to that seen by Topchy and Punch [32] who saw little difference between trees ‘fine-tuned’ at the end and their baseline GP performance. Along with one dataset (qsar-aquatic-toxicity) that exhibits no statistical difference, we have already suggested that tuning-at-end may cause overfitting for the dow-chemical dataset. It is tempting to observe that the mean model size after tuning-at-end is effectively the same as before tuning, and infer that over-parameterized models are being tuned to overfitting. However, like dow-chemical, tuning-at-end for the auto-mpg dataset also makes minimal change to the mean node counts but delivers a clear reduction in test MSE. Moreover, other datasets in Table 7 – most conspicuously, yacht-hydrodynamics – suggest that tuning-at-end can simultaneously increase the mean node count (i.e. select a more complex model) but decrease test MSE.
The comparable results incorporating standardization in (Table 8) reveal a much clearer picture than without standardization in Table 7). Here, tuning-at-end unambiguously reduces test MSE while increasing mean tree sizes (with the sole exception of yacht-hydrodynamics where the trees are the same sizes).
When viewing the results in Tables 7 and 8, it is important to bear in mind that the as-evolved populations from which the best performing trees are being selected are identical in each pair of comparisons – the only thing that is being changed by tuning is the values of the constants and hence the training MSE measures. Since we infer that Pareto-driven MOGP produces a spectrum of models ranging from small-and-underfitted to large-and-overfitted, there seems some variability in which sort of model is being promoted by tuning-at-end to be the best performing: sometimes what we presume the baseline GP is evolving as small, underfitted models are being tuned to be best. Other times, what we suspect are evolved as large, overfitted models are best performing after tuning. This behavior is, of course, in sharp contrast to tuning-during which consistently produces smaller trees. The above observations imply that large diversity in the final population is key so we conjecture that these effects would not be seen in single objective GP where model complexity is typically capped by a user-defined Koza-style hard depth limit. Overall, tuning-at-end would thus seem to warrant further focused research.
Tables 9 and 10 make the comparisons between tuning-during and tuning-at-end for unstandardized (Table 9) and standardized (Table 10) features, respectively. We can anticipate that, while tuning-at-end would certainly lead to improvements in test errors, but tuning-during would be superior since application of constant tuning at every stage in the evolution would have a more profound effect on guiding the evolutionary search process. It is obvious that the results with standardization are superior both in terms of test error and mean tree size.
4.3 Overall comparison
In this paper, we have considered a total of six different strategies: the baseline GP with and without standardization, and each of these with constant optimization both during and at the end of the evolution. It is interesting to ask which is the best overall technique? We can conveniently address this question using the Bayesian hierarchical test over multiple datasets due to Corani et al. [4],Footnote 5 this being the Bayesian equivalent of the frequentist signed-rank test usually recommended for such comparisons [6], but avoiding some of the questionable assumptions of the frequentist test, such as assuming all the error differences are i.i.d. across all datasets [4]. Comparison of the test MSE errors over all ten datasets are presented in Table 12: we present only results for MSE comparisons since these are of greatest interest. In order to present and discuss the results compactly in a table, we have adopted the abbreviations shown in Table 11 for the various methods.
Table 12 should be interpreted as the probability of the method in a given column in the topmost row being superior to the method listed in the leftmost column. So, to take the first row as an example, the probability that ‘SB’ is superior to ‘B’ over all ten datasets is 0.982. Similarly, the probability that ‘BE’ is superior to ‘BD’ is 0.314, namely ‘BE’ is actually inferior to BD’.
Determining an unambiguous overall ranking of methods from these pairwise tests is not straightforward since the measures in Table 12 are probabilities and not distances. Nonetheless, we can observe in general that:
-
‘B’, the baseline GP, is the worst performer of all, and is bettered by every other method.
-
‘SB’ appears to be second worst performer as it is bettered by every other method bar one (‘B’).
-
‘BE’ ranks as the third worst method.
-
‘BD’ and ‘SBD’ are equivalent performers over all datasets.
-
Given the choice between ‘SBD’ and ‘SBE’, ‘SBD appears superior.
It should be borne in mind, however, that the results in Table 12 are ‘aggregated’ over all ten datasets. So, for example, the above comments about ‘SBD’ appearing better than ‘SBE’ overall need to be tempered by observations over individual datasets. This simply reflects the limitations of statistical inference.
4.4 Algorithm run times
Clearly, the versions of the algorithm that employed numerical optimization alongside evolution will consume significantly more CPU time than the plain evolutionary algorithm. (To offset that, MSE performance is generally improved, of course.) The greatly increased run time for constant tuning was acknowledged several times by Kommenda et al. [17] although these authors did not directly quantify these increases on real instances of GP runs. Table 13 compares run timesFootnote 6 for the energy-efficiency-heat dataset since this is around the median run time across all datasets. Since optimizing execution times was not the principal objective of this work, the run times shown in Table 13 could almost certainly be reduced by careful tuning of the code, but that was outside the scope of the present study.
It is obvious from Table 13 that including constant tuning (either during evolution or only on the final population) significantly increases the run time over the baseline GP. We have used the steady-state evolutionary paradigm in this work which generates (and evaluates) two children at each breeding stage so the ‘tuning-during’ algorithm was able to employ two threads, each evaluating the fitness of a single offspring. The ‘tuning-at-end’ variant also used two threads for direct comparability, but could make much greater use of multithreading by using one thread to tune each individual in the final population; to repeat: our focus here has not been minimizing run time. It is also clear from Table 13 that there is significant variability in the run times as evidenced by the sizable standard deviations.
Over and above multithreading, the code implementation here used symbolic differentiation to evaluate derivatives [30] largely for convenience, and because optimizing code efficiency was not focus of this research. Aside from other refinements, initial code profiling suggest that a very large fraction of the total run time may be consumed in evaluating derivatives, which might be much more efficiently implemented, to the same precision, using automatic differentiation [1]. This is obviously an area for future work to facilitate more rapid turnaround of experiments.
The upper bound on the total number of iterations for ‘tuning-during‘ was 10,000 breeding operations \(\times\) 50 iterations per child = 500,000) whereas for ‘tuning-at-end’, the upper bound was \(100 \times 500 = 50,000\) iterations (a population of 100 \(\times\) 500 iterations). The constant optimizations for some trees required fewer than the limiting numbers of iterations – 50 for ‘tuning-during’ and 500 for ‘tuning-at-end’ – but many optimizations were terminated on exceeding these fixed iteration limits. Optimizations that exceeded their iteration limit displayed slow convergence, but provided approximate rather than exactly optimal solutions. Table 13 records the percentages of optimizations that exceeded their iteration limits (“% exceeded” column). For ‘tuning-during’, this was around 20% – in fact, the iteration limit of 50 was initially selected on the basis of about 80% of optimizations meeting their relative convergence limit of \(1 \times 10^{-4}\). It is also noteworthy that all the tuning-at-end optimizations converged within the limit of 500 iterations although only around 10% of trees had converged after the 50 iterations, the limit imposed on the tuning-during approach. The average tree sizes in the as-evolved population were, however, larger than those in the population generated with embedded optimization – see Table 4 for precise comparisons which probably explains i) the longer overall run times as evaluating a large tree obviously takes longer than evaluating a small tree, and ii) larger trees containing probably more constants on average require more iterations to converge.
Generally, iterative algorithms converge slowly [16] either because of the objective function being optimized is poorly scaled – not the case here – or because some of the parameters being optimized are ‘weakly’ coupled to the objective function. To illustrate this, in least-squares fitting of a straight line with a function \(y = (a+b)x + c\), calculating precise values of both a and b is indeterminate, but finding a single, precise value for the sum \((a+b)\) is feasible (debarring any other numerical difficulties). We have seen ample evidence of ‘double constant’ tree termination in [29] in a study of tree pruning. An obvious way to explore this issue (in future work) is to perform minor simplification of a tree by replacing all ‘double constants’ with a single constant using a simple rule-based substitution, which may remove numerical indeterminacy, and hopefully reduce slow convergence. Whether this significantly addresses the slow convergence problem remains to be seen since many other tree configurations involving unidentifiable constants are conceivable; potentially these could be diagnosed by a failure of some of the constant parameters to converge to stable values. Overall, more work remains to be done in the numerical optimization of GP trees.
Kommenda et al [17] have noted that nonlinear optimizations generally require an initial point within the ‘basin of attraction’ of the global minimum although do not appear to have addressed the point in practice. In a similar vein, we took the initial starting values of the optimization as being those constant values present in the as-bred child. A multistart algorithm [27, 28] could suppress possible convergence to local optima albeit at the cost of increased run time. Whether there is any benefit to this approach is an area for future work. (In passing, we note that the common practice – also used here – of starting GP runs with a number of different initial populations is actually an example of a multistart strategy.)
In terms of run time, the increasing availability of computing power makes constant tuning a practical proposition. Although the computational demands of tuning are significant, the computations are far from intractable; few would argue that very heavily compute-intensive deep learning is impractical because of its computing requirements. Indeed, the implementation of GP on the graphical processing units (GPUs) that facilitate much of deep learning has already received attention, e.g. [19].
Finally, the unexpectedly promising performance of tuning-at-end is worthy of more attention if only from a computational standpoint. For the steady-state evolutionary algorithm used in this paper, tuning-during allows only a limited amount of multithreading to reduce the run time: after each breeding stage, only two threads can be (straightforwardly) used, one to tune the constants in each child. Regardless of evolutionary strategy – steady-state or generational – tuning-at-end over a fixed population opens up the possibility of using one thread to optimize the constants in each individual in the final population thereby allowing a very much larger degree of multithreading, and thereby reduced run time. In an era of multicore processors, this could effectively address the computational issue with constant tuning.
4.5 Does constant tuning really improve performance?
In the results described up to this point, the numbers of evolutionary steps – that is, manipulations of the model structure – have been held constant at 10,000 between the no-tuning and with-tuning experiments; although we have employed a steady-state evolutionary approach throughout, for comparison, this is the equivalent of around 111 generations of a generational algorithm (assuming 10% elitism). (Convergence of a stochastic algorithm is always a problematic issue: the figure of 10,000 offspring was selected on the based on the observation that the rate of improvement appeared subjectively small after this number of iterations.) As discussed in Sect. 4.4, however, the run time with tuning is dominated by the numerical optimization of the constants due in significant part to the large number of tree evaluations required at each iteration of the SLSQP algorithm. We have limited the maximum number of SLSQP iterations to 50 (for tuning-during) although some optimizations converged in only 2-3 iterations. Across all datasets, it is difficult to be very precise about the total number of tree evaluations required due the large variability, but 200,000 is maybe a representative figure. It is interesting to pose the question of whether a budget of, say, 200,000 tree evaluations would be better spent solely on evolutionary optimization, and to ignore the complications of numerical optimizations? To address this question, we have run both the baseline and baseline-with-standardization algorithms to generate 200,000 offspring (as opposed to the 10,000 used hitherto) but without constant tuning, and compared the performance to the baseline-with-tuning (BD) and baseline-with-standardization-and-tuning (SBD), respectively that both used 10,000 evolutionary iterations. Running the evolutions for a very large number of iterations has produced slightly better-trained solutions albeit with a diminishing rate of success in finding these solutions.
The comparisons of results are included in Table 14 for the comparison without standardization, and Table 15 with standardization; the interpretation of these tables is identical to the previous such tables.
A number of points is apparent from Tables 14 and 15: firstly, the picture for individual dataset comparisons for test MSE indicates that sometimes the baseline (with or without standardization) for 200,000 tree evaluations performs compellingly better than the (S)BD method. Sometimes the reverse is true. For the baseline case, the split is 4-to-6 datasets better with just evolutionary search, and with standardization the split is 5-to-5.
The fact that for around half the datasets the better strategy is dispensing with numerical parameter tuning altogether (but still using very long run times) throws open the question of the value of numerical parameter calibration in GP, and whether, as is often believed, GP is not good at fine-tuning constant values. For some datasets it seems highly effective, for others the exact opposite is true. As a further confounding factor, predictor standardization appears to vary which datasets benefit from numerical parameter optimization.
Overall, the results in this section suggest that the dependence on dataset requires further research. This, in a sense, is disappointing as one of the hopes for GP is that can robustly automate the generation of predictive models without the need for the careful configuration typical of other methodologies. We speculate that one possible reason for the dependence on dataset could be that the ‘optimized’ models for different datasets have differing sensitivities to the precision of constant values due to the underlying properties of the data. If a dataset best matches a model with low sensitivities to constant precision then directing the tree evaluation budget to searching only over model structures is maybe the better strategy. If, on the other hand, the best models have high sensitivity to constant value precision then numerical tuning may be the better option. We stress this is speculation and would require further research, possibly exploring the sensitivities of ‘optimized’ models [31].
Turning to model sizes, an unambiguous trend emerges from Tables 14 and 15. The baseline GP – both with and without standardization – produces mean model sizes a factor 3-4 larger than with parameter calibration. The fact that tree sizes continue to grow with increasing numbers of evolutionary iterations while producing lower training errors implies that GP is able – at least in some cases – to effectively search for better constant values, presumably by synthesizing ever more complex trees. It is worth reiterating that the numbers of tree evaluations are roughly the same here so there is no overwhelming computational advantage in dispensing with numerical parameter tuning; the models produced by numerical tuning are, however, significantly smaller which is advantage for the practical deployment of GP. It is also noteworthy that the use of standardization yields larger models than without standardization in 8 out 10 datasets in Tables 14 and 15.
4.6 Statistical comparisons
The Bayesian comparison procedures developed by Benavoli et al. [2] were originally motivated to compute the posterior probabilities of performance differences between two classifiers. This formulation included the notion of a region of probable equivalence (ROPE) – a range around zero for which any difference between classifier performances could be regarded as so small as to be of no practical importance. Thus the ROPE quantifies the size effect of the difference in performance; in their work on classifiers, Benavoli et al. suggested a \(\pm 1\)% difference in classification accuracy as a possible figure although they recognized this would be context-dependent. Defining a ROPE allows direct calculation of the probability mass falling inside the ROPE – that is, the probability that there is no practical difference between the algorithms being compared. It is entirely possible for the performance of one method to appear statistically superior to another with, say, 99% probability, but for all of that probability mass to fall within the ROPE meaning that the difference between the methods is actually of no practical significance. In the present regression context, we have taken the ROPE to be of zero width because defining a range of MSE differences that are not practically significant is problematic for the datasets used here. Nonetheless, size effects are an important aspect of comparing regression models, and could, in principle, be incorporated by quantifying the variance of the measurements to be predicted: any test MSE error difference less than, say, \(2\times\) the noise variance of the experimentally measured predictors is very unlikely to be meaningful because the differences would be ‘in the noise’. Unfortunately, as far as we are aware, none of the standard datasets typically used in research studies on regression carries such information. Since benchmark datasets for GP have attracted attention in the past [33], we suggest the scope of benchmarks should be extended to explicitly address predictor noise and thereby the assessment of size effects.
5 Conclusions
In this paper we have extended previous analyses of constant tuning in genetic programming (GP) trees using gradient-descent style numerical algorithms to the steady-state multiobjective GP setting.
Along with considering the effects of constant tuning, we have also examined the effect of standardizing the dataset features. We find that, in general, standardization is beneficial in that it improves the test error performance of the models produced. We also conclude that standardization tends to produce larger models, seemingly in direct contradiction of recent work by Owen et al. [25] and Dick et al [7]. Further work is needed to understand these differences.
The effects of constant tuning during evolution – that is, optimizing the constant values for each child as soon as it is created – is always beneficial. It reduces both the test set error and the size of the best-generalizing trees when compared to the same method that omits constant optimization. This observation holds whether the features are standardized or not although standardization does generally appear to be of further benefit even for constant tuning in some cases.
We have also examined the effect of tuning the tree constants at the end of a conventional GP run. This turned out to be surprisingly effective in terms of improving generalization although the effects on tree size were rather mixed, and warrant further investigation.
Given a fixed computational budget of some number of tree evaluations, for around half the datasets it appears beneficial to run the GP evolution for longer rather than devote some of those tree evaluations to directly tuning the model constants; for the other half of the datasets, the exact opposite is true and parameter calibration yields superior test set errors. Using a long evolutionary run does, however, produce trees that are a factor of 3-4 larger than with numerical constant optimization. Clearly, this dependence on dataset requires further research.
In terms of statistical testing, we have employed a Bayesian procedure that avoids many of the objections to the null hypothesis statistical testing (NHST) traditionally used for performance comparison. This Bayesian test can explicitly incorporate a region of practical equivalence (ROPE) that allows future extension to considering the size effect between competing models; we point out, however, that existing benchmark datasets typically do not contain the necessary information on measurement noise to take this important step.
We have identified a number of areas for future work aimed at better understanding and developing constant tuning in GP models. In particular, the numerical aspects surrounding convergence of the optimization routines justifies additional attention.
Notes
In this work, we have used the implementation of the SLSQP optimizer from the NLOpt Version 2.6.2 library. See https://nlopt.readthedocs.io/en/latest/.
This is the classical maximum likelihood formulation.
We have used the Stan Bayesian inference engine version 2.26.1 (https://mc-stan.org/) together with the Stan model file provide by Corani and co-workers (https://github.com/BayesianTestsML/tutorial/tree/master/hierachical), and the default settings.
Evaluated on an HPC cluster (https://docs.hpc.shef.ac.uk/en/latest/sharc/cluster_specs.html#sharc-specs) comprising 2.40 GHz Intel Xeon E5-2630 v3 processors running Linux Centos 7.
References
A.G. Baydin, B.A. Pearlmutter, A.A. Radul, J.M. Siskind, Automatic differentiation in machine learning: a survey. J. Mach. Learn. Res. 18(1), 5595–5637 (2017)
A. Benavoli, G. Corani, J. Demšar, M. Zaffalon, Time for a change: a tutorial for comparing multiple classifiers through Bayesian analysis. J. Mach. Learn. Res. 18(77), 1–36 (2017)
G. Corani, A. Benavoli, A Bayesian approach for comparing cross-validated algorithms on multiple data sets. Mach. Learn. 100(2–3), 285–304 (2015). https://doi.org/10.1007/s10994-015-5486-z
G. Corani, A. Benavoli, J. Demšar, F. Mangili, M. Zaffalon, Statistical comparison of classifiers through Bayesian hierarchical modelling. Mach. Learn. 106(11), 1817–1837 (2017). https://doi.org/10.1007/s10994-017-5641-9
J. Demšar, Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 7, 1–30 (2006)
J. Demšar, On the appropriateness of statistical tests in machine learning. In: 3\(^{\rm rd}\) Workshop on Evaluation Methods for Machine Learning (ICML 2008). Helsinki, Finland (2008). http://www.site.uottawa.ca/ICML08WS/papers/J_Demsar.pdf
G. Dick, C.A. Owen, P.A. Whigham, Feature standardisation and coefficient optimisation for effective symbolic regression. In: Genetic and Evolutionary Computation Conference (GECCO ’20), pp. 306–314. Cancún, Mexico (2020). https://doi.org/10.1145/3377930.3390237
T. Dou, P. Rockett, Comparison of semantic-based local search methods for multiobjective genetic programming. Gen. Prog. Evol. Mach. 19(4), 535–563 (2018). https://doi.org/10.1007/s10710-018-9325-4
D. Dua, C. Graff, UCI Machine Learning Repository. University of California, School of Information and Computer Science, Irvine, CA (2019). http://archive.ics.uci.edu/ml
C. Fonseca, P.J. Fleming, Multiobjective optimization and multiple constraint handling with evolutionary algorithms - Part I: a unified formulation. IEEE Trans. Syst., Man Cybern. - Part A: Syst. Humans 28(1), 26–37 (1998). https://doi.org/10.1109/3468.650319
S. Geman, E. Bienenstock, R. Doursat, Neural networks and the bias/variance dilemma. Neural Comput. 4(1), 1–58 (1992). https://doi.org/10.1162/neco.1992.4.1.1
T. Hastie, R. Tibshirani, J. Friedman, The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer, New York, 2009)
M.T. Heath, Scientific Computing: An Introductory Survey (McGraw-Hill, New York, 2005)
F. Hutter, L. Kotthoff, J. Vanschoren (eds.), Automated Machine Learning: Methods, Systems, Challenges. Springer (2018). https://www.automl.org/wp-content/uploads/2019/05/AutoML_Book.pdf. In press, available at http://automl.org/book
M. Keijzer, Improving symbolic regression with interval arithmetic and linear scaling. In: C. Ryan, T. Soule, M. Keijzer, E. Tsang, R. Poli, E. Costa (eds.) European Conference on Genetic Programming (EuroGP 2003), pp. 70–82. Essex, UK (2003). https://doi.org/10.1007/3-540-36599-0_7
C.T. Kelley, Iterative Methods for Optimization. Society for Industrial and Applied Mathematics, Philadelphia, PA (1999). https://doi.org/10.1137/1.9781611970920
M. Kommenda, B. Burlacu, G. Kronberger, M. Affenzeller, Parameter identification for symbolic regression using nonlinear least squares. Gen. Prog. Evol. Mach. 21(3), 471–501 (2019). https://doi.org/10.1007/s10710-019-09371-3
D. Kraft, Algorithm 733: TOMP-Fortran modules for optimal control calculations. ACM Trans. Math. Softw. 20(3), 262–281 (1994). https://doi.org/10.1145/192115.192124
W.B. Langdon, Graphics processing units and genetic programming: an overview. Soft Comput. 15(8), 1657–1669 (2011). https://doi.org/10.1007/s00500-011-0695-2
C. Nadeau, Y. Bengio, Inference for the generalization error. Mach. Learn. 52(3), 239–281 (2003)
J. Ni, R.H. Drieberg, P.I. Rockett, The use of an analytic quotient operator in genetic programming. IEEE Trans. Evol. Comput. 17(1), 146–152 (2013). https://doi.org/10.1109/TEVC.2012.2195319
M. Nicolau, A. Agapitos, Choosing function sets with better generalisation performance for symbolic regression models. Gen. Program. Evol. Mach. (2020). https://doi.org/10.1007/s10710-020-09391-4
M. Nicolau, J. McDermott, Genetic programming symbolic regression: what is the prior on the prediction?, in Genetic Programming Theory and Practice XVII. ed. by W. Banzhaf, E. Goodman, L. Sheneman, L. Trujillo, B. Worzel (East Lansing, MI, 2019), pp. 201–225
R.S. Olson, J.H. Moore, TPOT: A Tree-Based Pipeline Optimization Tool for Automating Machine Learning (Springer International Publishing, Cham, 2019), pp. 151–160. https://doi.org/10.1007/978-3-030-05318-5_8
C.A. Owen, G. Dick, P.A. Whigham, Feature standardisation in symbolic regression, in AI 2018: Advances in Artificial Intelligence. ed. by T. Mitrovic, B. Xue, X. Li (Wellington, New Zealand, 2018), pp. 565–576. https://doi.org/10.1007/978-3-030-03991-2_52
R. Poli, W.B. Langdon, N.F. McPhee, A Field Guide to Genetic Programming. Published via http://lulu.com and freely available at http://www.gp-field-guide.org.uk (2008). http://dces.essex.ac.uk/staff/rpoli/gp-field-guide/A_Field_Guide_to_Genetic_Programming.pdf
A.H.G. Rinnooy Kan, G.T. Timmer, Stochastic global optimization methods Part I: clustering methods. Math. Program. 39(1), 27–56 (1987). https://doi.org/10.1007/BF02592070
A.H.G. Rinnooy Kan, G.T. Timmer, Stochastic global optimization methods Part II: Multi level methods. Math. Program. 39(1), 57–78 (1987). https://doi.org/10.1007/BF02592071
P. Rockett, Pruning of genetic programming trees using permutation tests. Evol. Intell. 13(4), 649–661 (2020). https://doi.org/10.1007/s12065-020-00379-8
P. Rockett, Y. Kaszubowski Lopes, T. Dou, E.A. Hathway, d(Tree)-by-dx: Automatic and exact differentiation of genetic programming trees. In: H.P. García, L. Sánchez-González, M.C. Limas, H. Quintián-Pardo, E.S.C. Rodríguez (eds.) 14\(^{\rm th}\) International Conference on Hybrid Artificial Intelligent Systems (HAIS2019), pp. 133–144. León, Spain (2019). https://doi.org/10.1007/978-3-030-29859-3_12
A. Saltelli, S. Tarantola, F. Campolongo, M. Ratto, Sensitivity Analysis in Practice: A Guide to Assessing Scientific Models (Wiley, Hoboken, 2004)
A. Topchy, W.F. Punch, Faster genetic programming based on local gradient search of numeric leaf values. In: L. Spector, E.D. Goodman, A. Wu, W.B. Langdon, H.M. Voigt, M. Gen, S. Sen, M. Dorigo, S. Pezeshk, M.H. Garzon, E. Burke (eds.) Genetic and Evolutionary Computation Conference (GECCO-2001), pp. 155–162. San Francisco, CA (2001). http://www.cs.bham.ac.uk/~wbl/biblio/gecco2001/d01.pdf
D.R. White, J. McDermott, M. Castelli, L. Manzoni, B.W. Goldman, G. Kronberger, W. Jaśkowski, U.M. O’Reilly, S. Luke, Better GP benchmarks: community survey results and proposals. Gen. Program. Evol. Mach. 14(1), 3–29 (2013). https://doi.org/10.1007/s10710-012-9177-2
Acknowledgements
The author is grateful to James McDermott for supplying the copy of the Dow Chemical dataset used in this work as well as for interesting discussions on testing methodologies. We are also grateful for the insightful comments and suggestions of a number of anonymous reviewers.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rockett, P. Constant optimization and feature standardization in multiobjective genetic programming. Genet Program Evolvable Mach 23, 37–69 (2022). https://doi.org/10.1007/s10710-021-09410-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10710-021-09410-y