Intelligent Data Analysis and Fusion Techniques in Pharmaceuticals, Bioprocessing and Process Control
B F Buxton, S B Holden and P C Treleaven
Department of Computer Science, University College London,
Gower Street, London, WC1E 6BT.
INTErSECT Faraday Partnership Flagship Project,
4 January 1999- 3 July 2002
| Grant Reference | GR/M43975 | Programme | MANUFACTURING ENGINEERING |
| Organisation | University College London | Scheme | INTErSECT Faraday |
1 Introduction and Background
This project was one of the INTErSECT Faraday Partnership’s four "flagship projects" announced in September 1998. Its aim was to develop hybrid intelligent systems for data mining and data fusion in the pharmaceutical and bioprocessing industries. Within the spirit of the Faraday Partnerships, the objective was not only to carry out high quality research, but also to create new and effective links between academe and industry. In the INTErSECT, Intelligent Sensors for Control Technologies, Faraday Partnership managed by NPL and Sira, the focus was similarly on creating links between academe and industry in the wider sensor, measurement, instrumentation and data analysis area with particular emphasis on the process and manufacturing industries. Other flagship projects were thus simultaneously launched by INTErSECT on the development of luminescent thermometry, acoustic emission sensing and signal diagnostics, and the application of data fusion to the analysis of combustion processes.
This project was funded by means of an EPSRC grant of just over £210,000 with additional substantial support in kind from the industrial collaborators: SmithKline Beecham, Unilever, Glaxo-Wellcome and SPSS (initially as Integral Solutions Limited). The grant, announced in September 1998, provided direct support for a postdoctoral research fellow and a PhD student but, in addition, three Sira/UCL Postgraduate Training Partnership PhD students were initially linked to the project, one with support from Zeneca. The plan was for the four PhD students to work one each closely with each of the four major industrial collaborators: SmithKline Beecham, Unilever, Zeneca and Glaxo-Wellcome. SmithKline Beecham and Glaxo-Wellcome merged during the course of the project to become Glaxo-SmithKline, but arrangements for the students were left unchanged by the merger. Zeneca, in contrast, split into AstraZeneca and Syngenta and, although the project maintained links to both companies, the main contact was at Syngenta. Two of the Sira students, working with SmithKline Beecham and Unilever, commenced in October 1998. A third began study in January 1999 at the same time as the directly supported EPSRC student, whilst a research assistant was appointed at the beginning of April 1999.
The intention of the project was that each of the students would study the development and application of modern machine learning and data mining techniques appropriate to the particular problems of their industrial collaborators, whilst the research fellow would develop and implement the system for data fusion and combining techniques. The latter was to include the combination of methods routinely used in the industry and in other applications and, time allowing, methods developed during the course of the project. Each of the four industrial ‘users’ posed a data mining application of particular interest to them: Unilever the analysis of food preference data which typically have many features and few records; SmithKline Beecham, the analysis of high throughput screening data often with large, unbalanced data sets; Glaxo-Wellcome the use of structure activity relationships in the early stages of drug discovery, and Syngenta/AstraZeneca applications in process optimisation, often involving the need to store and analyse data collected over the duration of production batches.
The state of the art in these industrial application areas was typically represented by the use of conventional statistical regression and fitting techniques and by the use of well-established machine learning techniques such as linear classifiers, multi-layer perceptron (MLPs) and radial basis function (RBF) neural networks and decision trees. These are, often implemented by use of industry standard data mining tools such as the Clementine system developed by ISL but, by the time the project commenced, marketed by SPSS. Clementine, made available to the academics as part of SPSS’s contribution to the project, was thus an invaluable resource, used for rapid, state of the art, data mining work throughout much of the project.
When the project was proposed, we thus considered that the area was ripe: (a) for the application of more sophisticated, modern data-mining and machine learning techniques, and (b) the introduction of some kind of hybrid intelligent system methodology that could be used to combine the results obtained with a variety of methods. In particular, the latter approach had been very successful at UCL in application to the finance industry where function replacing and function combining hybrid systems had been developed [1] and had led, in due course, to the establishment of Searchspace, a very successful company specialising in the area. In addition, it was evident at the time of the proposal that the support vector machines and other large margin methods were showing a great deal of promise in the literature, both in terms of their theoretical advances and their performance reported on publicly available, benchmark data sets [2,3]. Finally, there was progress in developing a deeper understanding of multiclassifier systems and it was beginning to emerge as to: (i) how to characterise their performance [4], and (ii) how effectively to combine a variety of systems [5].
We were thus in the fortunate position of having a number of research opportunities to pursue with the scope for doctorate study and system development. Each of which was likely to lead to academic advances. At the same time, we could collaborate with well-motivated industrial partners with real data available and with real interest in novel solutions and their application. This, which would force us to address issues that might otherwise be overlooked in curiosity driven, pure research.
2 Key Advances and Supporting Methodology
Of the four research studentships associated with the project, three, including that directly supported by the EPSRC grant award, were successful, and have resulted in one thesis submitted and the viva completed [6], a second thesis has just been submitted [7], and one is nearing completion [8]. The first student has continued on a research project at UCL funded by GSK [9]. The second one worked for a year at Imperial College CSTM, before joining Credit Suisse First Boston this autumn. Finally, the third one worked as a teaching systems developer in the Department of Computer Science at UCL, prior to taking up at research post on a recently announced Wellcome grant in collaboration with colleagues in the Wolfson Institute for Biomedical Research [10]. Between them, the work of these three research students has, to date, led to six publications as detailed in the IGR form, with more likely to be produced. The fourth studentship, unfortunately, was not successful. The first student initially appointed resigned within twelve months of commencement. The second, replacement student, after successful initial work arising out of early undergraduate project work under the supervision of Sean Holden that led to a publication given in the IGR form, also resigned and eventually took up a research position at Cambridge University.
The particular advances made by the successful students were first, by David Corney, in the modelling of the food preference data work in collaboration with Unilever. This, which led to development of: a semi-supervised feature selection algorithm, a semi-supervised ensemble method for regression, a clustering evaluation technique, and an outlier detection technique for clustering. This was essentially a modelling problem in which feature selection is essential because of the lack of preference data, as noted above, which is difficult and expensive to obtain. Overall, machine learning and modelling techniques such as Bayes networks show performance similar to traditional statistical methods with small improvements in accuracy in some cases. However, in general, the machine learning techniques are dependent on fewer assumptions than the statistical methods and are more robust, giving more reliable results should particular assumptions not be borne out in practice. Details are given in Corney’s thesis [6] and in the two papers produced.
The second PTP student, Robert Burbidge, whose thesis was recently submitted, currently on the point of submission, worked on the development of heuristic methods for support vector machines (SVMs) with applications to drug discovery initially in collaboration, with SmithKline Beecham. Preliminary work [11] on public cheminformatic data established that SVMs could, with little user effort, match or outperform the best, capacity controlled neural network, even if the latter were carefully hand-tuned. Since SVMs are protected against the adverse effects of high-dimensional representations and are robust with respect to measurement inaccuracies, they appear to be ideally suited to the analysis of screening data. However, owing to the implicit nature of the model generated by an SVM, it may, on such data, produce models that are very dense with the decision hyperplane represented by expansion on a large subset of compounds. This hinders model interpretability and increases the prediction computational time required for prediction on new, previously unseen data. This is, an important consideration as when it may be necessary to subject many millions of compounds to a virtual, electronic "pre-screen" and predictions must not be slow, in comparison say, to those that could be obtained by use of a neural network. A heuristic (STAR) was therefore developed for reducing the model complexity of an SVM during training which does not require pre- or post-processing of the data. Training time is also an important consideration because of the way the quadratic program at the heart of the SVM learning algorithm scales with size of the training data sets used. Other heuristics were thus developed for on-line parameter tuning during training of an SVM and for early stopping of the quadratic program based on the predicted performance of the model. Details are given in Burbidge’s thesis draft [7] and in the papers published.
The work on application of SVMs described above mostly used an RBF kernel. The third student, Matthew Trotter, working initially in collaboration with Glaxo-Wellcome, focussed on the design of SVMs specifically tailored to the drug discovery environment. This was motivated by comparative studies which showed that SVMs considerably outperformed a range of other supervised machine learning techniques on a variety of real pharmaceutical data. A nearest-neighbour/SVM hybrid was found capable of improving performance by taking account of local effects within the data. This work raised the classification accuracy of simple, polynomial kernels to equal and sometimes better that of a heuristically tuned radial basis function (RBF) kernel. Further benefits of the hybrid were the provision of a free parameter more intuitive to the non-expert user than the RBF width and the ability to flag outliers, itself an important consideration in drug discovery. Further consideration of local effects and the needs of non-expert users lead to the development of an SVM kernel with no free parameters, using a generic data representation and associated similarity measure. The new kernel provided an easily understandable transformation of the data and greatly reduced the time required for parameter selection, while maintaining a level of accuracy comparable to that of conventional SVM kernels with tuned parameters. Details are again to be given in Trotter’s thesis under preparation [8] and in the papers published and further papers currently under preparation.
3 Project Plan Review: Key Advances in Data and Decision Fusion
The work of the three successful PhD students described above proceeded to plan. The work of the research fellow and the key advances made in this part of the project are discussed under this section as there was a change of staff midway through the project and, with it, a slight change of plan and a rather greater change of emphasis.
The first research assistant, Appolo Tankeh, appointed in March 1999 prior to submission of his thesis at Imperial College, worked on the project for twelve months during which the focus was on developing an integrated formulation capable of dealing, on the same footing, with the recognition of patterns within temporal, process data, and within static data. The starting point for this work was the discrete non-linear regression formulation of Zhu and Billings [12] which can be related to techniques for assessing data and model complexity and to neural networks, RBFs, fuzzy logic and rule induction system approaches (see for example [13, 14]). The latter suggested a possible relationship with the kind of hybrid intelligent systems used in earlier work in finance [1], but we were unable to pursue this further or complete the work for publication as Appolo Tankeh left for a post with IBM in the USA in March 2000.
This meant that valuable time was lost and that the data fusion system work was in danger of falling behind the work of the PhD students. To avoid this, it was decided to adopt a more direct approach and immediately proceed to building a system that could combine the outputs of a wide variety of classifier systems. As noted in the introduction, by 2000, research on multiclassifier systems was very topical (see for example the reviews articles in [15]) and many approaches to classifier fusion were being studied, theoretically, experimentally and in applications. In particular, it was well appreciated that there are three fundamental reasons for combining classifiers [16]: statistical because the amount of training data is limited and almost always too small compared to the size of the hypothesis space; computational because, even given enough data, many learning algorithms find only locally optimal solutions; and representational because it may not be possible to represent the true decision function in the chosen hypothesis space. It was appreciated that multiple classifier systems could, in principle alleviate or solve these problems [16], but there was no way, in general, of generating appropriate classifier combinations. All three problems occur in non-trivial applications such as those of interest here. In particular, in drug discovery, the class boundaries may be complicated in the Kolmogorov sense (see for example [17]). This is sometimes paraphrased as the classes being "long and thin in the feature space" which means, for example, that very many isotropic RBFs may be needed in an SVM kernel, if that approach is being used.
There are two broad approaches to classifier fusion, known as decision optimisation and coverage optimisation [17]. Coverage optimisation assumes a fixed combination rule and manipulates the classifiers as in boosting [3], whilst decision optimisation assumes a given set of classifiers and manipulates the combination rule. Some work on boosting was carried out by Sean Holden and two of the research students (Matthew Trotter and, briefly, by Jeevani Wickramaratna – see publication listed on the IGR form), but it was found to be is difficult to obtain predictable results with strong initial classifiers. Decision optimisation was therefore selected for study, since it was known that fixed combination rules (such as: sum, max, min, median, and majority voting) though they can be very effective, are not usually optimal. Furthermore, it is straightforward to develop a set of suitable classifiers by use of data mining tools such as Clementine. Fixed combination rules (such as: sum, max, min, median, and majority voting), though they can be very effective, are not usually optimal.
Three things then needed to be brought together in order to optimise the combination of a given ensemble of classifiers: (i) a means of generating a sufficiently general, but capacity controlled combination function (the danger of statistical over-fitting problems are is ever present!); (ii) a suitable measure of classifier performance, and (iii)
characterisation of a classifier’s output so that an appropriate intelligent combination function can be constructed. It is difficult, for example, to put a value on the cost of a missed drug discovery (false negative), even though the costs of false positives, which do not turn out to be useful leads, can be quantified. A robust measure of classifier performance is particularly important. For example, it is difficult to put a value on the cost of a missed drug discovery (false negative), even though the costs of false positives, which do not turn out to be useful leads, can be quantified.was therefore needed. Fortunately, the area under the ROC (receiver-operating characteristic (ROC) curve provides just such a well-defined statistical measure, viz: an estimate of the Wilcoxon statistic. Moreover, it was shown [44] how this measure could be used to combine classifiers so as to construct a Maximum Realisable ROC. However, this construction uses only the decisions output by the members of an ensemble of binary classifiers. Given, for example, a confidence measure on their outputs, our hypothesis was: (a) that it should be possible to construct useful combination functions giving superior performance and, (b) that genetic programming (GP) with suitable capacity control (ie. protection against "bloat" in the GP) would be an effective way of sampling the important areas of the hypothesis space and thus of generating effective classifier combinations.This has been completely borne out in practice. All that was required was to find a genetic programming expert who would have the skill to build the requisite system and to carry out the required experiments. We were very fortunate to recruit W B Langdon in May 2000. He quickly took charge of this research agenda, showed that GP could indeed be used in this way, demonstrated the method on several publicly available datasets as illustrated below, and embarked on a close collaboration with Dr Steven Barrett of SmithKline Beecham on applying the approach to drug discovery data mining problems. Dr Barrett, now of GSK, was also the industrial collaborator for Robert Burbidge’s PhD work.

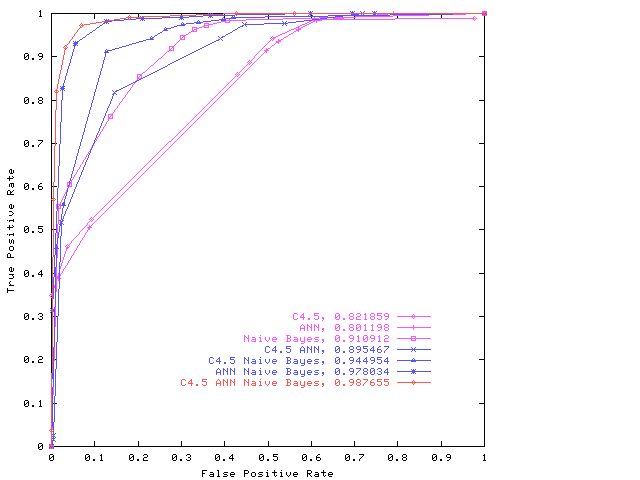
ROC for the combined classifier system produced by the GP on the Grey Landsat data. For simplicity only the convex hull or maximum realisable ROC [4] is given for each combination. See W B Langdon and B F Buxton in MCS 2001 for details.
Six papers have been published on this method of classifier combination as listed in the form. The early papers illustrated the approach using publicly available benchmark applications, many of which were used in [4], whilst later papers address applications in drug discovery. More detailed experiments and evaluations are underway in collaboration with GSK on the current RAIS project [GR/S03546/01]. This includes a new comparison with the application of boosting techniques, a blind trial and consideration of other applications in addition to drug discovery. In the original proposal, it was envisaged that the systems would be developed and tested on drug discovery/screening problems and the approach evaluated by application to problems in other domains, for example, on corrosion data available from NPL. In practice, the system was developed and tested on a variety of public datasets, then applied to problems in drug discovery for the fusion of a variety of classifier systems. The extent of the development work carried out is such that there was thus little need and, unfortunately, no time for the proposed work on other datasets within the scope of the Faraday project.
In addition, several other papers were produced as also listed in the IGR form, addressing issues such as bloat, convergence, program size and function distribution, and application of GP data mining methods in other areas, most notably text classification. As mentioned in the introduction, one of the graduate students from the project, David Corney, is continuing work on text data mining with support from GSK [9].
4 Research Impact and Benefits to Society
As detailed in the IGR form, the project has led to five journal publications and sixteen conference publications. Project investigators (BFB and SBH) have given several seminars at universities in the UK whilst the research fellow (WBL) has given several invited talks at conferences abroad. In addition, in collaboration with Dr Riccardo Poli (University of Birmingham), the research fellow has completed a book on the "Foundations of Genetic Programming", published recently by Springer-Verlag.
The subsequent RAIS project on the application of genetic programming to data fusion and data mining in the pharmaceutical industries [GR/S03546/01] commenced in September 2002. This project, also in collaboration with Dr Steven Barrett of GSK, is designed to enable the work to be further evaluated in the drug discovery area at GSK by means of more detailed, more extensive trials, and application to other targets, and also for its potential in other data mining areas in pharmaceuticals research to be explored.
In addition, with the assistance of Dr Suran Goonatilake, one of the founders of Searchspace, and Professor David Jones, head of our new Bioinformatics Unit, we have been exploring the potential for setting up a company. Plans have been discussed with our GSK collaborators and discussions have taken place with three venture capitalists: MVM Limited, Atlas Venture and the Wellcome Development fund administered by Catalyst. Research to identify the best opportunities for such a company and selection of the optimal first target areas for commercial work is being carried out with the assistance and advice of leading biological and medical researchers within UCL and the Bloomsbury area. In the current financial climate, only a very modest operation would be feasible in the near term, but we are confident that there are viable opportunities for such a company.
Finally, this project and our established track record in the development and application of intelligent systems in the financial and retail areas gave us a platform from which, with the assistance of staff from the Gatsby Institute for Computational Neuroscience and Professor David Jones, and the support of many of our industrial collaborators, the Department could launch a new masters programme in Intelligent Systems. An MTP was thus sought and secured [18]. The programme commenced in September 2001 and the first cohort of 6 students have completed their studies. The programme commenced in September 2001 and the first cohort of 6 students have completed their studies. This was a modest beginning, but growth this year has been quite spectacular with 19 students currently enrolled.
5 Explanation of Expenditure
There have been no significant variations in the planned expenditure and all the major industrial collaborators have contributed as planned. The only deviation was that the delay in appointing the first research assistant and a brief (unpaid) sabbatical visit to Sweden for two months by the second research fellow meant that there were sufficient funds available to extend the period of research by three months to 3rd July 2002. Throughout the project, the enthusiastic efforts and support of staff at Unilever, SmithKline Beecham and Glaxo-Wellcome (now GSK), SPSS and Sira are all gratefully acknowledged. In particular, the three Sira UCL PTP students linked to the project enabled us to establish immediately a useful sized research team and to ensure that individual researchers could focus on collaborating effectively with one each of the industrial users. Sira also gave invaluable assistance in the supervision and training of the three PTP students, each of whom worked on-site at Sira as well as at their industrial collaborators for extended periods of study and research.
6 Further Research and Dissemination Activities
Research and development work is continuing in collaboration with GSK under the RAIS project as noted in section 3. On completion of the RAIS project we hope, in addition to the text mining project supported by GSK, to continue this work: (i) via an Engineering Doctorate project on improving the visualisation, user interface and application of GP to data mining in collaboration with GSK; and (ii) by seeking a platform grant to continue and extend the development, application and understanding of genetic and evolutionary programming techniques.
The decision to apply our intelligent systems approach to problems in the pharmaceutical and process industries has also proved very timely. Thus, it provided an important stepping-stone to establishment of one of the Joint Research Council Bioinformatics Units in the Department of Computer Science at UCL [GR/R47455/01] which has now led to fully fledged collaboration with many of the biological scientists at UCL and elsewhere, including the Wolfson Institute for Biomedical Research [10] and the department of Biochemistry and Molecular Biology, the Institute of Child Health, the Institute of Molecular Pathology and the European Bioinformatics Institute [19]. Finally, a 150 processor Beowulf machine was obtained under the Strategic Research Initiative for use on computationally intensive problems in bioinformatics [20] in collaboration with the department of Biochemistry and Molecular Biology at UCL. Spare capacity on this machine has also been used latterly for our ongoing classifier combination work.
In addition to the dissemination opportunities
provided by the INTErSECT Partnership and publication and presentation
of our results as detailed on the IRG form, members of the project
have been very active in giving seminars and invited talks, both
within the UK and internationally, as noted in section 4 above. Wider
dissemination has included attendance at many of our regular project
meetings by representatives from other INTErSECT flagship projects,
visits and presentations to other INTErSECT partners at Rolls-Royce
(who were not participants of this project) and at the University of
Manchester, a presentation on the INTErSECT collaborative experience
to the DTI, two presentations at NCAF meetings, presentations at EPSRC
workshops on natural computation and applications of genetic
programming, and two articles for dissemination to a wider, scientific
and industrial audience, in Measurement and Control. One of the latter
articles on our data/decision fusion work received the Scientific
Instrument Makers’ Annual Award best paper prize for 2001. As much
information about the project as the confidential requirements of our
collaborators permits, including a
tutorial introduction to our
data/decision fusion work, is available on our
Referencesweb pages at UCL.
[1] S Goonatilake and P Treleaven, (Eds), "Intelligent Systems for Finance and Business", John Wiley and Sons, 1995.
[2] V N Vapnik, "The Nature of Statistical Learning Theory", Springer-Verlag, New York, 1995.
[3] Y Freund and R E Schapire, "Experiments with a new boosting algorithm", in Proc. 13th International Conference on machine Learning, Bari, Italy, 1996, pp. 148-156, 1996.
[4] M J J Scott, M Niranjan and R W Praeger, "Realisable classifiers: Improving operating performance on variable cost problems", in Proc. of 9th British Machine Vision Conf, Southampton, 14-17 Sept 1998, edited by Lewis PH & Nixon MS, vol 1, pp.305-315, 1998.Scott Niranjan et al
[5] J Kittler, M Hatel, R P W Duin, and J Matas, "On combining classifiers", IEEE PAMI, 20(3), 226-239,1998.Kittler et al 1998.
[6] D Corney, "Intelligent Analysis of Small Data Sets for Food Design", PhD Thesis, Department of Computer Science, UCL, 2002.
[7] R Burbidge, "Heuristic Methods for Support Vector Machines with Applications to Drug Discovery", PhD thesis submitted, Department of Computer Science, UCL, 2002.
[8] M W B Trotter, "The Application of Support Vector Machines to Drug Discovery", in preparation, PhD Thesis, Department of Computer Science, UCL.
[9] D T Jones (PI) and B F Buxton, research funded by GSK (G2558, overall value £143, 984) in November 2001.
[10] D H Beach et al, "Functional genomics of pluripotent stem cells and their progeny", Wellcome grant (overall value £5,119,123) announced in February 2002.] WIBR
[11] R Burbidge, M W B Trotter, S Holden and B F Buxton, "Drug Design by Machine Learning: SVMs for Pharmaceutical Data Analysis", presented at the AISB'00 Symposium on artificial Intelligence in Bioinformatics, April, 2001; printed in Computers & Chemistry, Vol 26(1), pp. 5-14, 2001. JCIC Cheminformatics and AISB
[12] Q M Zhu and S A & Billings, "Fast orthogonal identification of nonlinear stochastic models and radial basis function neural networks", Int. J. of Control, 64, pp871-886, 1996.
[13] Q M Zhu and S A Billings, "Parameter estimation for stochastic nonlinear models", Int. J. of Control, 57, pp 309-333, 19934.?
[14] L X Wang and J M Mendel, "Fuzzy basis functions, universal approximation, and orthogonal least square learning", IEEE Trans. Neural Networks, 3, pp 807-814, 1992.Wang and Mendel 1992.
[15] J Kittler and F Roli, (eds), MCS 2000, LNCS 1857, Springer-Verlag, Berlin Heidelberg, 2000.MCS 2000
[16] T G Dietterich, "Ensemble methods in machine learning", in MCS 2000 edited by J Kittler & F Roli, LNCS 1857, pp 1-15, Springer-Verlag, Berlin Heidelberg, 2000. [15]
[17] T K Ho, "Complexity of classification problems and comparative advantages of combined classifiers", in MCS 2000 edited by J Kittler & F Roli, LNCS 1857, pp 1-15, Springer-Verlag, Berlin Heidelberg, 2000.
[18] S B Holden (PI), B Buxton and P C Treleaven, "MTP in Intelligent Systems", GR /30217, £565778 EPSRC funding, £827908 total support, April 2001.
[19] C Orengo et al, "Integrating transcriptomics and structural data to reveal protein functions", Wellcome grant (overall value ~£800,000) announced August 2002.
[20] B F Buxton, S A Sorensen (PI) and J M Thornton, "Multiprocesor System for Computational Bioinformatics", part of funding awarded to UCL under the Strategic Equipment Initiative, £167650, SEI0015, Sept. 2000-March 2002.
BFB 24 Oct 2002. Conversion to HTML WBL 21 Nov 2002. (Links updated 17 Oct 2008)