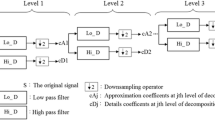
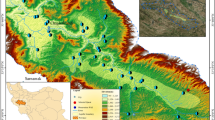
Abstract
Water constitutes an essential part of the earth as it helps in making the environment greener and support life. But water quality and availability are drastically affected by rising water pollution and its poor sanitation. Water gets contaminated due to the excessive use of chemicals by the industries, fertilizers, and pesticides by the farmers. Not only the surface water, groundwater and river water are also getting contaminated. Several published work in Indian context have used different models for the prediction of water quality. Some of them performed poorly due to the presence of irrelevant and missing data in the training samples. Moreover, these studies have assessed water quality on the basis of biochemical oxygen demand (BOD) and coliform and chemical oxygen demand (COD), whereas dissolved oxygen(DO) is one of the most important parameters in terms of water quality assessment as it is considered a key determinant of pollution. Thus, there is a strong need to categorically identify and visualize the DO as one of the key components responsible for deteriorating the quality of water in Indian context. The main objective of this work is to build a wavelet genetic programming (WGP)-based workflow model for the assessment of water quality in 13 rivers of Uttar Pradesh region. WGP model has a unique feature of discarding the redundant and irrelevant data values from the source data. The proposed WGP model has given promising results which can be attributed to two factors: firstly, the novel use of Morlet wavelet in place of the widely popular Db wavelet, as the mother wavelet, and secondly, the use of MICE technique for missing value imputation in the pre-processing stage. The proposed model not only cleans the data but also demonstrates the feasibility of using DO values as one of the prime factors to assess the water quality.






Similar content being viewed by others
Data availability
River water data was taken from the UP-Pollution Control Board website titled “Water Quality Data of the Polluted River Stretches” available at http://www.uppcb.com/water-quality-data-stretches.htm for the years 2020 and 2021.
References
Altunkaynak, A., & Nigussie, T. A. (2017). Monthly water consumption prediction using season algorithm and wavelet transform–based models. Journal of Water Resources Planning and Management, 143. https://doi.org/10.1061/(ASCE)WR.1943-5452.0000761
Azad, A. S., Sokkalingam, R., Daud, H., Adhikary, S. K., Khurshid, H., Mazlan, S. N. A., & Rabbani, M. B. A. (2022). Water level prediction through hybrid SARIMA and ANN models based on time series analysis: Red Hills Reservoir case study. Sustainability, 14, 1843. https://doi.org/10.3390/su14031843
Azari, S. M., Bozorg-Haddad, O., & Loaiciga, H. (2020). State-of-art of genetic programming applications in water-resources systems analysis. Environmental Monitoring and Assessment, 192. https://doi.org/10.1007/s10661-019-8040-9
Baigang, D., Zhou, Q., Guo, J., Guo, S., & Wang, L. (2021). Deep learning with long short-term memory neural networks combining wavelet transform and principal component analysis for daily urban water demand forecasting. Expert Systems with Applications, 171. https://doi.org/10.1016/j.eswa.2021.114571
Emadi, A., Sobhani, R., Ahmadi, H., Boroomandnia, A., Zamanzad-Ghavidel, S., & Azamathulla, H. M. (2022). Multivariate modeling of agricultural river water abstraction via novel integrated-wavelet methods in various climatic conditions. Environment, Development and Sustainability, 24, 4845–4871. https://doi.org/10.1007/s10668-021-01637-0
Gao, A., Wu, S., Zhu, S., & Xu, Z. (2018). Wavelet and statistical analyses of river water quality parameters: A case study in the Lower Minnesota River. Water Practice and Technology, 13(4), 922–931. https://doi.org/10.2166/wpt.2018.101
Ghorbani, M. A., Khatibi, R., Mehr, A. D., & Asadi, H. (2018). Chaos-based multigene genetic programming: A new hybrid strategy for river flow forecasting. Journal of Hydrology, 562, 455–467. https://doi.org/10.1016/j.jhydrol.2018.04.054
Hadi, S. J., & Tombul, M. (2018). Monthly streamflow forecasting using continuous wavelet and multi-gene genetic programming combination. Journal of Hydrology, 561, 64–687. https://doi.org/10.1016/j.jhydrol.2018.04.03
Huan, J., Li, M., Xu, X., Zhang, H., Yang, B., Jianming, J., & Shi, B. (2022). Multi-step prediction of dissolved oxygen in rivers based on random forest missing value imputation and attention mechanism coupled with recurrent neural network. Water Supply, 22(5), 5480–5493. https://doi.org/10.2166/ws.2022.154
Huang, M., Tian, D., Liu, H., Zhang, C., Yi, X., Cai, J., Ruan, J., Zhang, T., Kong, S., & Ying, G. (2018). A hybrid fuzzy wavelet neural network model with self-adapted fuzzy c-means clustering and genetic algorithm for water quality prediction in rivers. Complexity, 2018, 8241342. https://doi.org/10.1155/2018/8241342
Jafari, H., Rajaee, T., & Kisi, O. (2020). Improved water quality prediction with hybrid wavelet-genetic programming model and Shannon entropy. Natural Resources Research, 29, 3819–3840. https://doi.org/10.1007/s11053-020-09702-7
Jamei, M., Ahmadianfar, I., Xuefeng, C., & Yaseen, Z. M. (2020). Prediction of surface water total dissolved solids using hybridized wavelet-multigene genetic programming: New approach. Journal of Hydrology, 589. https://doi.org/10.1016/j.jhydrol.2020.125335
Kumar, M., & Sahay, R. R. (2018). Wavelet-genetic programming conjunction model for flood forecasting in rivers. Hydrology Research, 49(6), 1880–1889. https://doi.org/10.2166/nh.2018.183
Lovrinovic, I., Srzić, V., Matić, I., & Brkić, M. (2022). Combined multilevel monitoring and wavelet transform analysis approach for the inspection of ground and surface water dynamics in shallow coastal aquifer. Water, 14(4), 656. https://doi.org/10.3390/w14040656
Liu, J., Ding, J., Ge, X., & Wang, J. (2021). Evaluation of total nitrogen in water via airborne hyperspectral data: Potential of fractional order discretization algorithm and discrete wavelet transform analysis. Remote Sens., 13(22), 4643. https://doi.org/10.3390/rs13224643
Marcelino, C. G., Leite, G. M. C., Celes, P., & Pedreira, C. E. (2022). Missing data analysis in regression. Applied Artificial Intelligence, 36(1). https://doi.org/10.1080/08839514.2022.2032925
Martínez-Acosta, L., Medrano-Barboza, J. P., López-Ramos, A., López, J. F. R., & López-Lambraño, A. A. (2020). SARIMA approach to generating synthetic monthly rainfall in the Sinú river watershed in Colombia. Atmosphere, 11(6), 602. https://doi.org/10.3390/atmos11060602
Mehr, A. D. (2021). Seasonal rainfall hindcasting using ensemble multi-stage genetic programming. Theoretical and Applied Climatology, 143, 461–472. https://doi.org/10.1007/s00704-020-03438-3
Mehr, A. D., & Gandomi, A. H. (2021). MSGP-LASSO: An improved multi-stage genetic programming model for streamflow prediction. Information Sciences, 561, 181–195. https://doi.org/10.1016/j.ins.2021.02.011
Mirzaei, A., Carter, S. R., Patanwala, A. E., & Schneider, C. R. (2021). Missing data in surveys: Key concepts, approaches, and applications. Research in Social and Administrative Pharmacy, 18(2), 2308–2316. https://doi.org/10.1016/j.sapharm.2021.03.009
Momeni, H., & Bonakdari, H. (2016). Forecasting monthly inflow with an extreme seasonal variation using the hybrid SARIMA-ANN model. Stoch Environ Res Risk Assess, 31, 1997–2010. https://doi.org/10.1007/s00477-016-1273-z
Poul, A. K., Shourian, M., & Ebrahimi, H. (2019). A comparative study of MLR, KNN, ANN and ANFIS models with wavelet transform in monthly stream flow prediction. Water Resources Management, 33, 2907–2923. https://doi.org/10.1007/s11269-019-02273-0
Raja, P. S., & Thangavel, K. (2019). Missing value imputation using unsupervised machine learning techniques. Soft Computing, 24, 4361–4392. https://doi.org/10.1007/s00500-019-04199-6
Rajaee, T., Khani, S., & Ravansalar, M. (2020). Artificial intelligence-based single and hybrid models for prediction of water quality in rivers: A review. Chemometrics and Intelligent Laboratory Systems, 200. https://doi.org/10.1016/j.chemolab.2020.103978
Ravansalar, M., Rajaee, T., & Kisi, O. (2017). Wavelet-linear genetic programming: A new approach modeling monthly streamflow. Journal of Hydrology, 549, 461–475. https://doi.org/10.1016/j.jhydrol.2017.04.018
Seo, Y., Choi, Y., & Choi, J. (2017). River stage modeling by combining maximal overlap discrete wavelet transform, support vector machines and genetic algorithm. Water, 9(7), 525. https://doi.org/10.3390/w9070525
Tadesse, K. B., & Dinka, M. O. (2017). Application of SARIMA model to forecasting monthly flows in Waterval River, South Africa. Journal of Water and Land Development., 35, 229–236. https://doi.org/10.1515/gold-2017-0088
Tripathi, A. K., Saini, H., & Rathee, G. (2022). Futuristic prediction of missing value imputation methods using extended ANN. International Journal of Business Analytics, 9(3). https://doi.org/10.4018/IJBAN.292055
Wang, X., Tian, W., & Liao, Z. (2020). Statistical comparison between SARIMA and ANN’s performance for surface water quality time series prediction. Environmental Science and Pollution Research, 28, 33531–33544. https://doi.org/10.1007/s11356-021-13086-3
Wu, J., & Wang, Z. (2022). A hybrid model for water quality prediction based on an artificial neural network, wavelet transform, and long short-term memory. Water, 14, 610. https://doi.org/10.3390/w14040610
Author information
Authors and Affiliations
Contributions
Mansi Gaonkar: data curation, investigation and analysis, writing—original draft, and visualization. Bhawna Saxena: writing—review and editing. Sandeep Kumar Singh: writing—review and editing, and supervision.
Corresponding author
Ethics declarations
Ethics approval
All authors have read, understood, and have complied as applicable with the statement on “Ethical responsibilities of Authors” as found in the Instructions for Authors and are aware that with minor exceptions, no changes can be made to authorship once the paper is submitted.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
S. No | Abbreviation | Full form |
|---|---|---|
1 | W-MGGP | Wavelet pre-processing with multi-gene genetic programming |
2 | WGEP | Wavelet gene expression programming |
3 | DWT | Discrete wavelet transform |
4 | LPM | Linear probability model |
5 | MGGP | Multi-gene genetic programming |
6 | PSR | Predictive state representation |
7 | FNN | False nearest neighbor |
8 | LASSO | Least absolute shrinkage and selection operator |
9 | GEP | Genetic evolutionary programming |
10 | FLGGP | Fixed-length gene genetic programming |
11 | LGP | Linear genetic programming |
12 | GP | Genetic programming |
13 | ARIMA | Autoregressive integrated moving average |
14 | WGP | Wavelet genetic programming |
15 | MSGP | Multi-stage genetic programming |
16 | GBT | Gradient boosting |
17 | ANN | Artificial neural networks |
18 | WANN | Wavelet artificial neural networks |
19 | ANFIS | Adaptive neuro-fuzzy inference system |
20 | WANFIS | Wavelet adaptive neuro-fuzzy inference system |
21 | FL | Fuzzy logic |
22 | SVM | Support vector machine |
23 | NF | Neuro-fuzzy |
24 | GA-NN | Genetic algorithm neural networks |
25 | WNN | Wavelet neural network |
26 | WSVR | Wavelet support vector regression |
27 | WLGP | Wavelet linear genetic programming |
28 | PCA | Principle component analysis |
29 | LSTM | Long short-term memory |
30 | RNNs | Recurrent neural networks |
31 | BP | Back propagation |
32 | SVR | Support vector regression |
33 | MSA | Multiple sequence alignments |
34 | MLP | Multilayer perceptron |
35 | MLR | Multiple linear regression |
36 | KNN | K-nearest neighbor |
37 | MODWT | Maximal overlap discrete wavelet transform |
38 | FWNN | Wavelet-based fuzzy neural network |
39 | CWT | Continuous wavelet transform |
40 | DDM | Data-driven modeling |
41 | NAR | Non-linear autoregressive neural network |
42 | RF | Random forest |
43 | ABM | Agent-based modeling |
44 | GBU | Gradient boosting unit |
45 | EMSP | Effective maliciousness score of permission |
46 | GBT | Gradient boosting technique |
47 | OAM | Open application model |
48 | LEM | Learnable evolution model |
49 | EP | Evolutionary programming |
50 | AR | Augmented reality |
51 | WNF | Wavelet neuro-fuzzy |
52 | WTC | Wavelet task clustering |
53 | SARIMA | Seasonal autoregressive integrated moving average |
54 | FNN | Feedforward neural network |
55 | RMSE | Root mean square error |
56 | NSE | Nash–Sutcliffe efficiency |
57 | MAE | Mean absolute error |
58 | R2 | Coefficient of determination |
59 | MAPE | Mean absolute percentage error |
60 | MSE | Mean square error |
61 | AICc | Akaike information criterion |
62 | AARE | Average absolute relative error |
63 | MARE | Mean absolute relative error |
64 | AOI | Index of agreement |
65 | CE | Coefficient of efficiency |
66 | NSC | Nearest shrunken centroid |
67 | SSE | Sum of squared errors |
68 | MPE | Mean prediction error |
69 | R | Coefficient of correlation |
70 | RE | Relative error |
71 | MRSE | Mean root squared error |
72 | PARE | Pooled average relative underestimation and over estimation errors |
73 | NDEI | Non-dimensional error index |
74 | Ai | Accuracy factor |
75 | P | Pearson correlation coefficient |
76 | bias | Absolute value of the average forecast error |
77 | d | Willmott index of agreement |
78 | pMAPE | Mean absolute percentage errors of peaks |
79 | EVS | Explain variance score |
80 | RPD | Residual prediction deviation |
81 | Dr | Discrepancy ratio |
82 | MSRE | Mean square relative error |
83 | MS4E | Mean higher order error |
84 | SI | Scatter index |
85 | MS | Mean squared error |
86 | SBC | Schwarz Bayesian criterion |
87 | DO | Dissolved oxygen |
88 | BOD | Biochemical oxygen demand |
89 | EC | Electrical conductivity |
90 | COD | Chemical oxygen demand |
91 | TDS | Total dissolved solids |
92 | TP | Total phosphorus |
93 | TKN | Total Kjeldahl nitrogen |
94 | NH3-N | Ammoniacal nitrogen |
95 | RD | River depth |
96 | RW | River width |
97 | CA | Cultivated area |
98 | OA | Orchard area |
99 | SSL | Suspended sediment load |
100 | SSC | Suspended sediment concentration |
101 | NO3+ | Nitrate |
102 | NH4+ | Ammonium |
103 | PO4 | Phosphate |
104 | TSS | Total suspended solids |
105 | NH4+-N | Ammonium nitrogen |
106 | WT | Water temperature |
107 | TU | Turbidity |
108 | db | Daubechies4 |
109 | dmey | Discrete Meyer |
110 | bior | Biorthogonal |
111 | coif | Coiflets |
112 | sym | Symlets (symmetrical wavelets) |
113 | MCAR | Missing completely at random |
114 | MAR | Missing at random |
115 | MNAR | Missing not at random |
116 | MICE | Multiple imputation by chained equations |
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Saxena, B., Gaonkar, M. & Singh, S.K. Study of the effectiveness of wavelet genetic programming model for water quality analysis in the Uttar Pradesh region. Environ Monit Assess 195, 1010 (2023). https://doi.org/10.1007/s10661-023-11489-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10661-023-11489-y